Trimmer-2 cDNA normalization kit
본문
Trimmer-2 cDNA normalization kit
- Rapid and reliable way to remove repeated transcripts from cDNA library
- Equalization of full-length-enriched cDNA before library cloning
- Simple procedure, no physical separation steps
- Fully compatible with Illumina / Solexa, ABI / SOLiD and Roche / 454 sequencing protocols
In an eukaryotic cell, the mRNA population constitutes approximately 1% of total RNA with the number of transcripts varying from several thousand to several tens of thousands. Normally, the high abundance transcripts (several thousand mRNA copies per cell) of as few as 5-10 genes account for 20% of the cellular mRNA. The intermediate abundance transcripts (several hundred copies per cell) of 500-2000 genes constitute about 40-60% of the cellular mRNA. The remaining 20-40% of mRNA is represented by rare transcripts (from one to several dozen mRNA copies per cell) [Alberts et al., 1994]. Such an enormous difference in abundance complicates large-scale transcriptome analysis, which results in recurrent sequencing of more abundant cDNAs.
cDNA normalization decreases the prevalence of high abundance transcripts and equalizes transcript concentrations in a cDNA sample, thereby dramatically increasing the efficiency of sequencing and rare gene discovery.
Trimmer-2 kit is designed to normalize amplified full-length-enriched cDNA prepared using Evrogen Mint cDNA synthesis kits. The resulting cDNA contains equalized abundance of different transcripts and can be used for construction of cDNA libraries and for direct sequencing, including high-throughput sequencing on the next generation sequencing platforms (Roche/454, ABI/SOLiD or Illumina/Solexa). The kit also includes special adapters allowing use of Clontech SMART-based kits for construction of cDNA intended for Trimmer-2 normalization.
 |
 |
 |
|---|
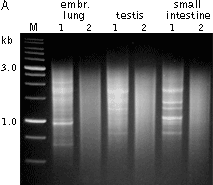
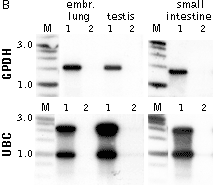
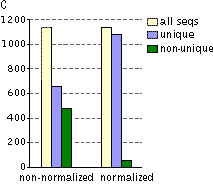
Typical cDNA normalization result.
(A) Agarose/EtBr gel-electrophoresis of non-normalized (1) and normalized (2) human cDNA samples; (B) concentration of abundant transcripts in these samples revealed by Virtual Northern blot. GPDH – glyceraldehyde-3-phosphate dehydrogenase; UBC – ubiquitin C; M, 1-kb DNA size markers (SibEnzyme). (C) Normalization significantly increases gene discovery rate of cDNA library. Transcript distribution in standard and normalized cDNA libraries from Aplysia neurons: yellow – all sequences, blue – unique sequences; green – non-unique sequences.
댓글목록
등록된 댓글이 없습니다.